
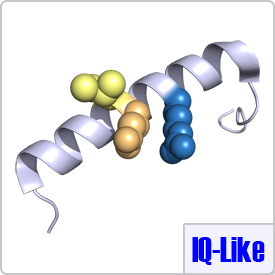
IQ-Like motifs are considered incomplete as they lack a second basic residue and final anchor (shown in pink) in the motif:[FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY]. CaM binding to these motifs is typically calcium-dependent. The IQ-Like motif is commonly found in ion channels. When binding to IQ-like motifs, CaM can either completely wrap around the target peptide as seen with the voltage-dependent L-type calcium channel (PDB: 2BE6) or remain in a semi-open conformation as seen with voltage-gated sodium channels (PDB: 2L53).
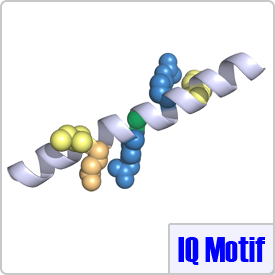
The complete IQ motif has a consensus sequence of: [FILV]Qxxx[RK]Gxxx[RK]xx[FILVWY] in which the residues in brackets can substitute for each other. CaM typically binds to the IQ motif in the absence of calcium. The model example for this group is myosin V (PDB: 2IX7).
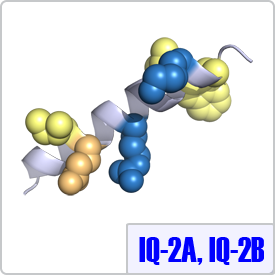
IQ-2A and IQ-2B were characterized from the second light chain of the myosin family of proteins. In these motifs, the spacing between the basic residues is different and the final anchor is a tryptophan instead of any hydrophobic residues. Thus, the consensus sequence for these motifs are: [ILV]QxxxRxxxx[LV][KR]xW and [IL]QxxCxxxxKxRxW, respectively. Currently, structures of these moitfs bound to calmodulin have not been determined. Thus, the IQ-2A peptide shown on the right is depicted on a model helix.
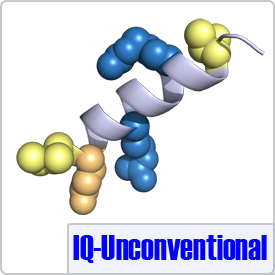
The unconventional motif was determined from sequence alignment of neuromodulin, neurogranin, and unconventional myosins. The only difference between the IQ motif and an IQ-Unconventional motif is the absence of a glycine at position 7 (shown in pink), which has little effect on the conformation of CaM when bound to its peptide targets (PDB: 4E53):[FILV]Qxxx[RK]xxxx[RK]xx[FILVWY].
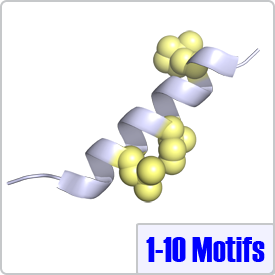
In this motif, two bulky hydrophobic residues are spaced by 8 amino acids: [FILVW]xxxxxxxx[FILVW]. Targets containing an extra anchoring residue at position 5 are called 1-5-10 motifs. In addition, 1-5-10 motifs preceded by basic residues are called basic 1-5-10 motifs. These motifs are found in CaM-dependent protein kinases (PDBs: 1CDM and 3GP2) as well as the pain receptor TRPV1 (PDB 3SUI). A 1-5-10 peptide is shown on the right.
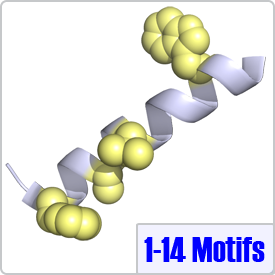
One of the most common motifs, 1-14 motifs like 1-10 motifs contain two bulky hydrophobic residues [FILVW], but are spaced by 12 amino acids. Targets may contain additional internal anchoring residues [FAILVW] at positions 5 and 8. Similarly to 1-5-10 motifs, 1-8-14 motifs preceded by basic residues are called "basic" 1-8-14 motifs. These motifs are found in enzymes such as nitric oxide synthase (PDB: 2LL6) as well as ion transporters such as: the plasma membrane calcium pump (PDB: 2KNE) and the cyclic nucleotide-activated olfactory channel (PDB: 1SY9). A 1-5-8-14 motif utilizing an alanine residue (position 5) is shown on the right.
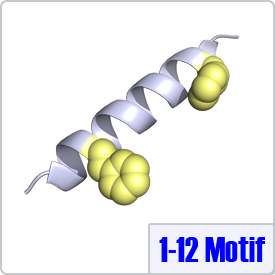
The Ikura lab proposed the 1-12 motif after an extensive literature search and analysis of confirmed CaM target proteins. Similar to the 1-10 and 1-14 class, 1-12 motifs contain two bulky hydrophic residues spaced 10 amino acids apart: [FILVW]xxxxxxxxxx[FILVW]. Although this motif has been identified in multiple enzymes, to date, there are no crystal structures of CaM bound to this motif deposited in the Protein Data Bank. The pepitide structure shown on the right depicts the two hydrophobic residues on a model helix.
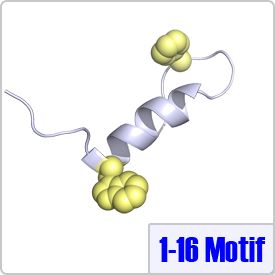
The 1-16 motif describes targets that contain two bulky hydrophobic residues spaced 14 residues apart: [FILVW]xxxxxxxxxxxxxx[FILVW]. CaM takes on a unique conformation when binding to the 1-16 motif. Typically, CaM binds to binds to 1-16 in a reverse orientation (the N-terminus of the peptide binds to the N-terminal domain of CaM) as observed in the crystal structure with CaM-dependent kinase kinase (PDB: 1CKK).

